Why we need LinkSet?
Introduction
LinkSet draws significant inspiration from InteractionSet, an excellent package developed by Aaron Lun. InteractionSet provides robust functionality for storing and manipulating genomic interaction datasets, offering powerful features like efficient subsetting operations and comprehensive GRanges methods.
While working extensively with enhancer-promoter interaction analyses, including Promoter Capture Hi-C (PC-HiC) datasets and enhancer-promoter inference in multiomic studies, I identified several limitations in InteractionSet that prompted the development of LinkSet:
- Enhancer-promoter interactions have an inherent directional and causal nature, where enhancers specifically regulate promoters in a unidirectional fashion. The InteractionSet framework, however, lacks explicit support for representing these crucial directional relationships.
- Although InteractionSet provides fundamental statistical capabilities like distance calculations, it offers limited analytical functionality for interaction data analysis. Additionally, it lacks comprehensive visualization methods that are essential for interpreting complex interaction datasets.
- The package has restricted options for file format conversions, making it challenging to work with diverse data formats commonly used in genomic interaction studies.
What linkSet can offer?
Direction and causality
LinkSet has two major features : Bait and Other End (OE). The Bait is the target region of interaction, for example, promoters in E-P interaction and bait in PC-HiC. The Other End is regulated region, for example, enhancers in E-P interaction.
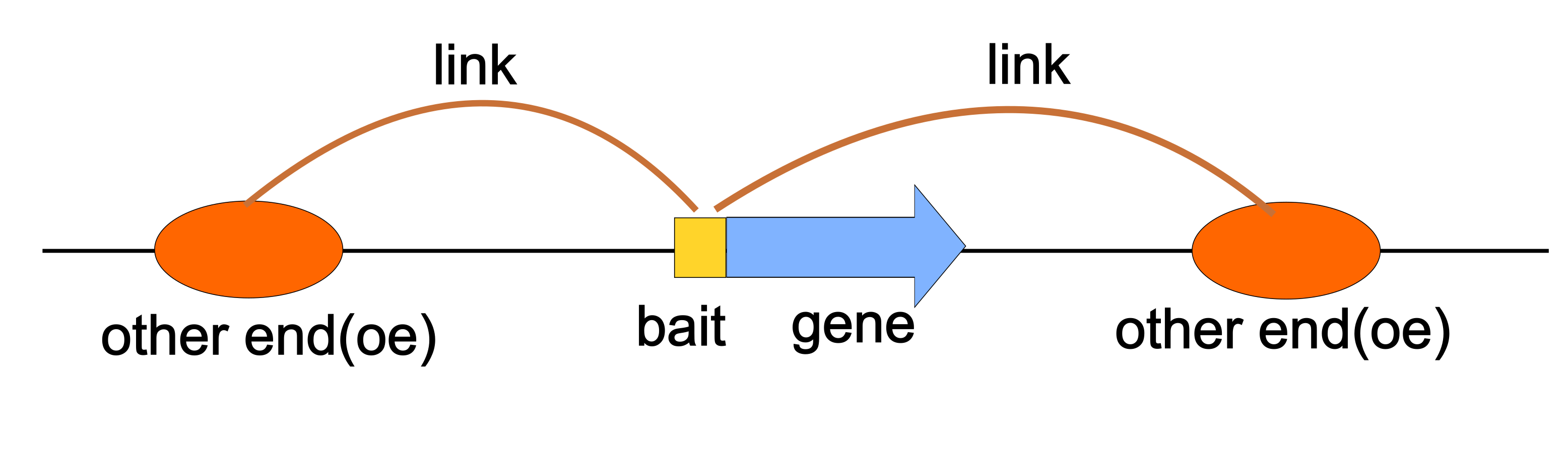
The Bait can be GRanges object, and it also can be string, such as Sp7 or ENSG00000170374. This is useful in enhancer-promoter inference methods in multiomic studies, like SCENT, as you won’t have the genomic coordinates of bait.
Statistical analysis
LinkSet provides basic diagnostic plots, such as distance distribution, and trans/cis ratio.
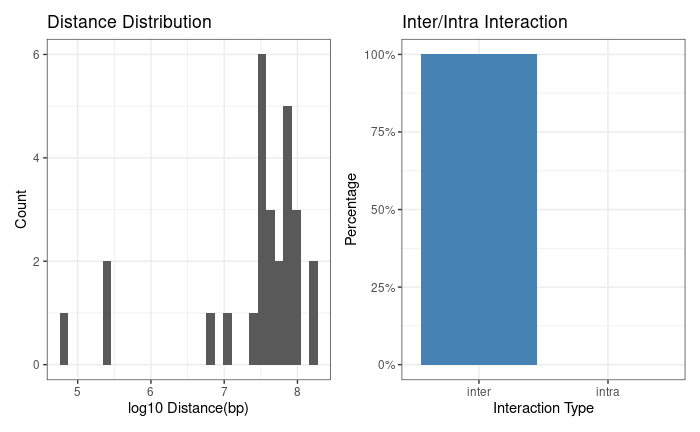
Also, you can reduce the regions and count the interaction frequency in the reduced regions. LinkSet provides methods to calculate the enhancers that regulate multiple promoters, and provide basic visualization. LinkSet provides basic diagnostic plots, such as distance distribution, and trans/cis ratio.
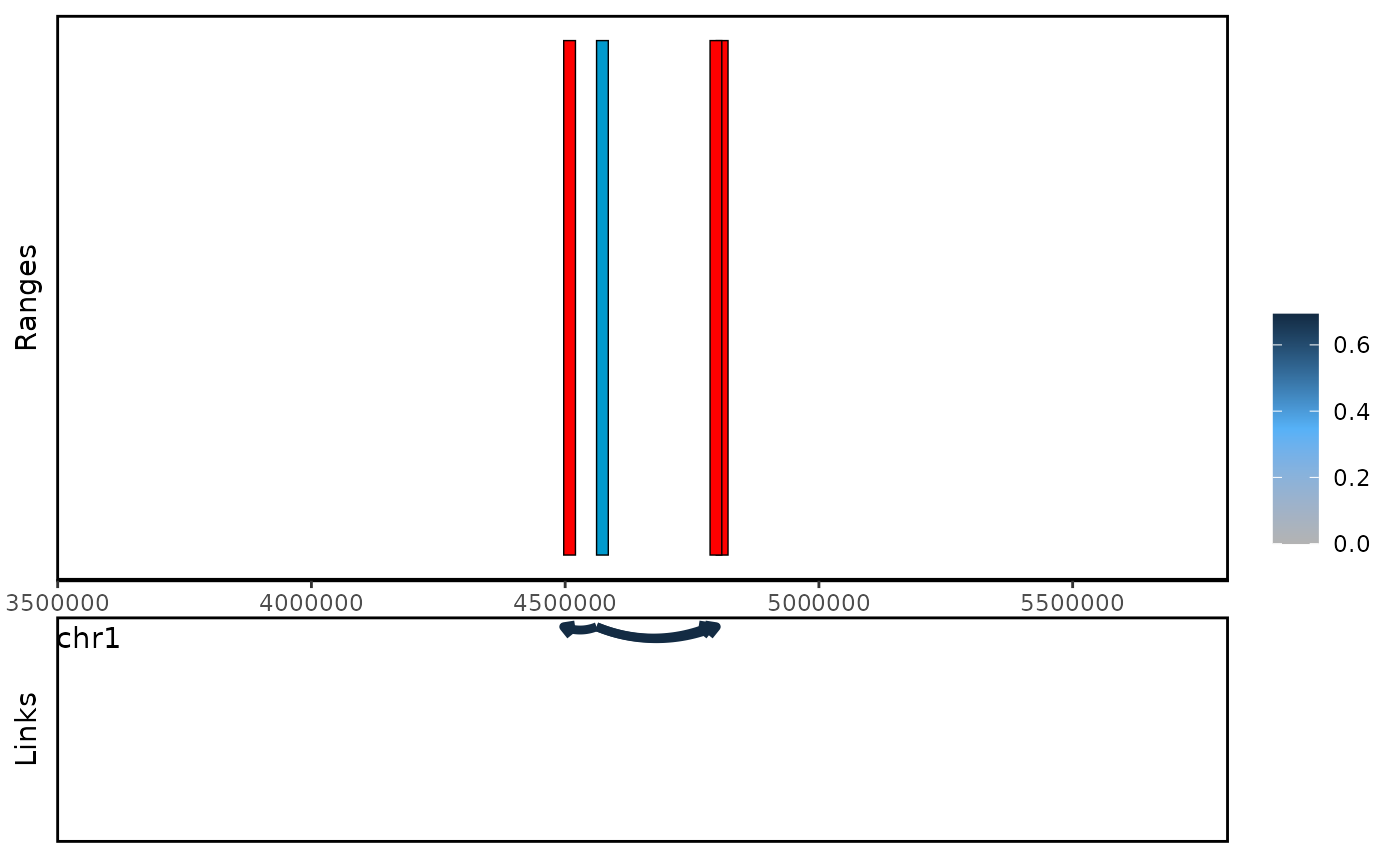
While the current visualization capabilities are still in development, I am actively working on enhancing the aesthetic appeal and functionality of these plots in upcoming releases. Future improvements will focus on creating more polished and publication-ready visualizations.
And I also implement the method CHiCANE to identify the high-confidence interactions, see run_chicane. However, it should be noted that CHiCANE was designed for PC-HiC data, and the result format is not suitable for the analysis of other types of interaction data.
File format conversion
LinkSet provides convenient methods to convert the interaction data between different formats. For example, CHiCAGO format in PC-HiC analysis, and GenomicInteractions format.
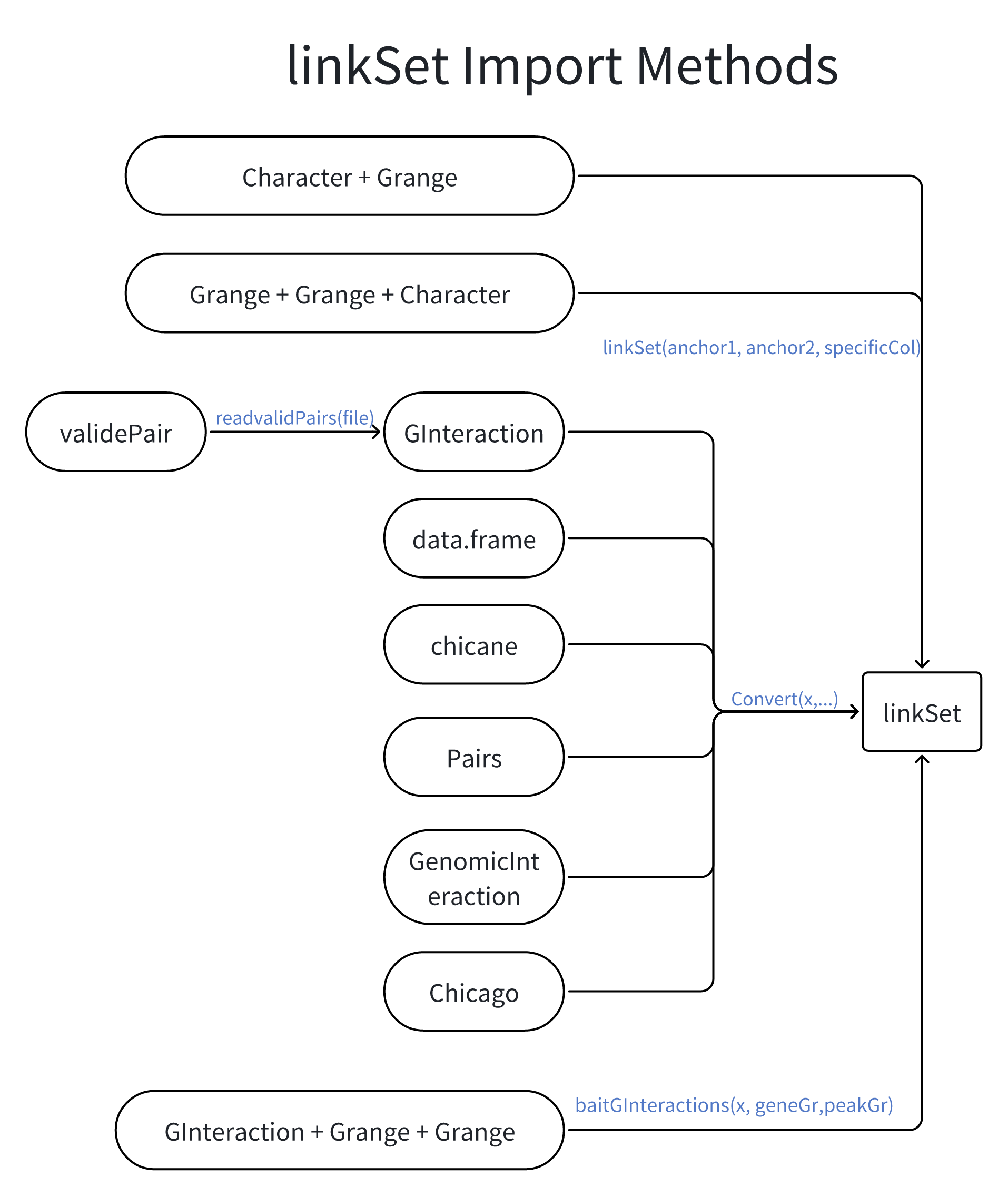
And it’s also easy to export the interaction data to other formats, such as ibed, WashU, etc.
Conclusion
LinkSet have been avaliable on github, and recently, I’m going to submit it to Bioconductor.
I welcome feedback and contributions from users to help shape the future development of linkSet and make it an even more valuable resource for genomic interaction analysis.
Enjoy Reading This Article?
Here are some more articles you might like to read next: