TrajAtlas
Trajectory Framework for Large-Scale Atlas
TrajAtlas was my first project, and it holds a special place in my heart. It came into my life during the hardest time, giving me the goal to walk out from the depression. Through this project, I gained a comprehensive understanding of academic research, learning how to identify a starting point, deliver presentations, and engage in academic writing. While I acknowledge that the project may not be perfect, I can confidently say it represents my best effort. I believe TrajAtlas contributes to the advancement of life sciences—perhaps a small step for humanity, but a significant leap for me personally. I am deeply grateful to everyone who supported me, especially Huan Liu, whose assistance was invaluable in bringing this paper to fruition.
The motivation for this project stems from the observation that osteoblasts have diverse origins. This idea was excellently reviewed by Noriaki Ono, who I was fortunate to have as one of my reviewers. I soon realized that there was no systematic analysis from a single-cell RNA perspective. While there has been a significant growth in scRNA datasets from various tissues, I believe I can utilize these datasets to construct an atlas to reveal the origins of osteoblast cells.
Then comes the Differentiation Atlas, the first atlas that focus on differentiation. The preparation process is long and difficult. I have no ideas whether datasets from different tissues can be integrate together. So I carefully collected the metadata, made lots of benchmarking, and I even pre-construct three tissue atlas seperately.
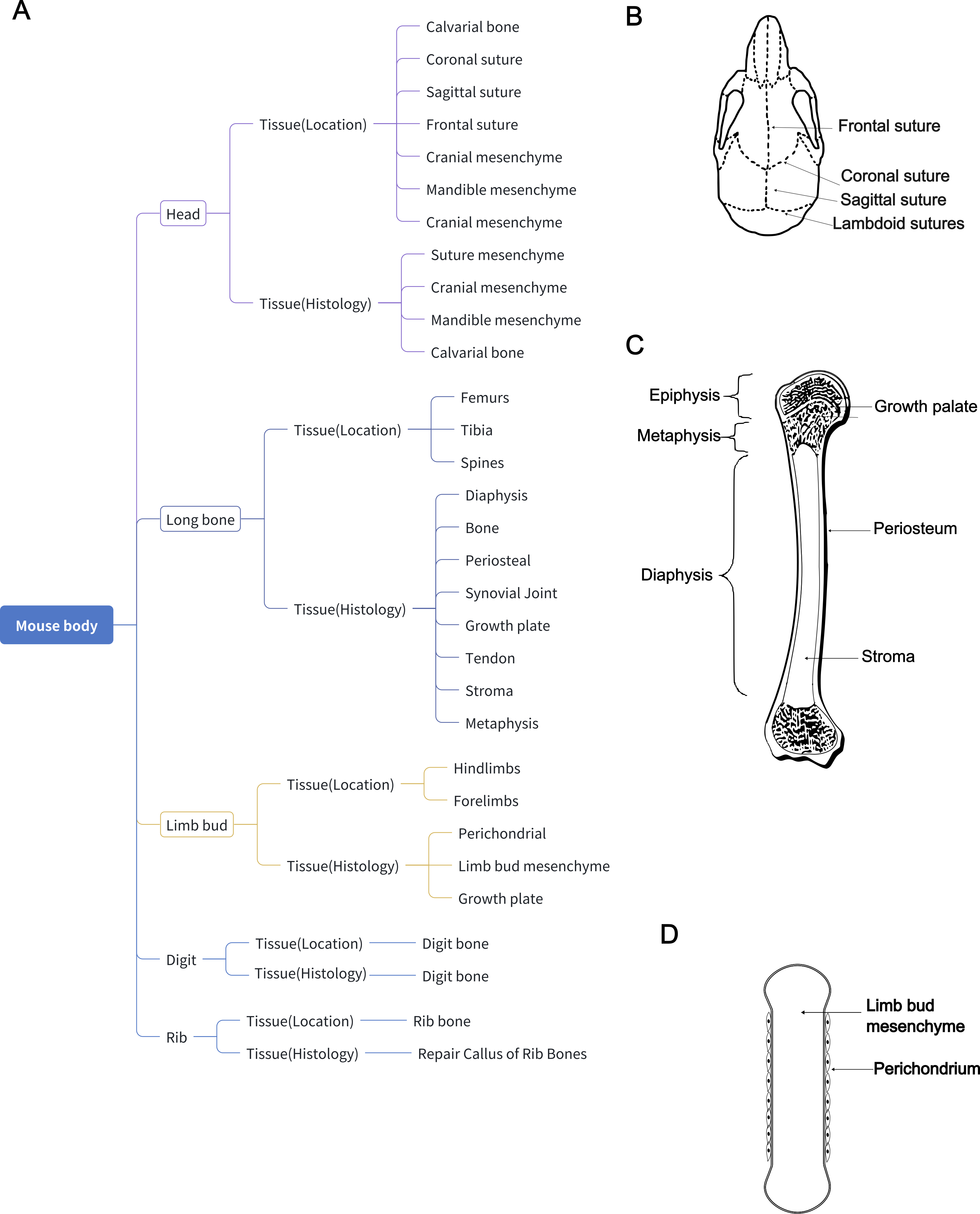
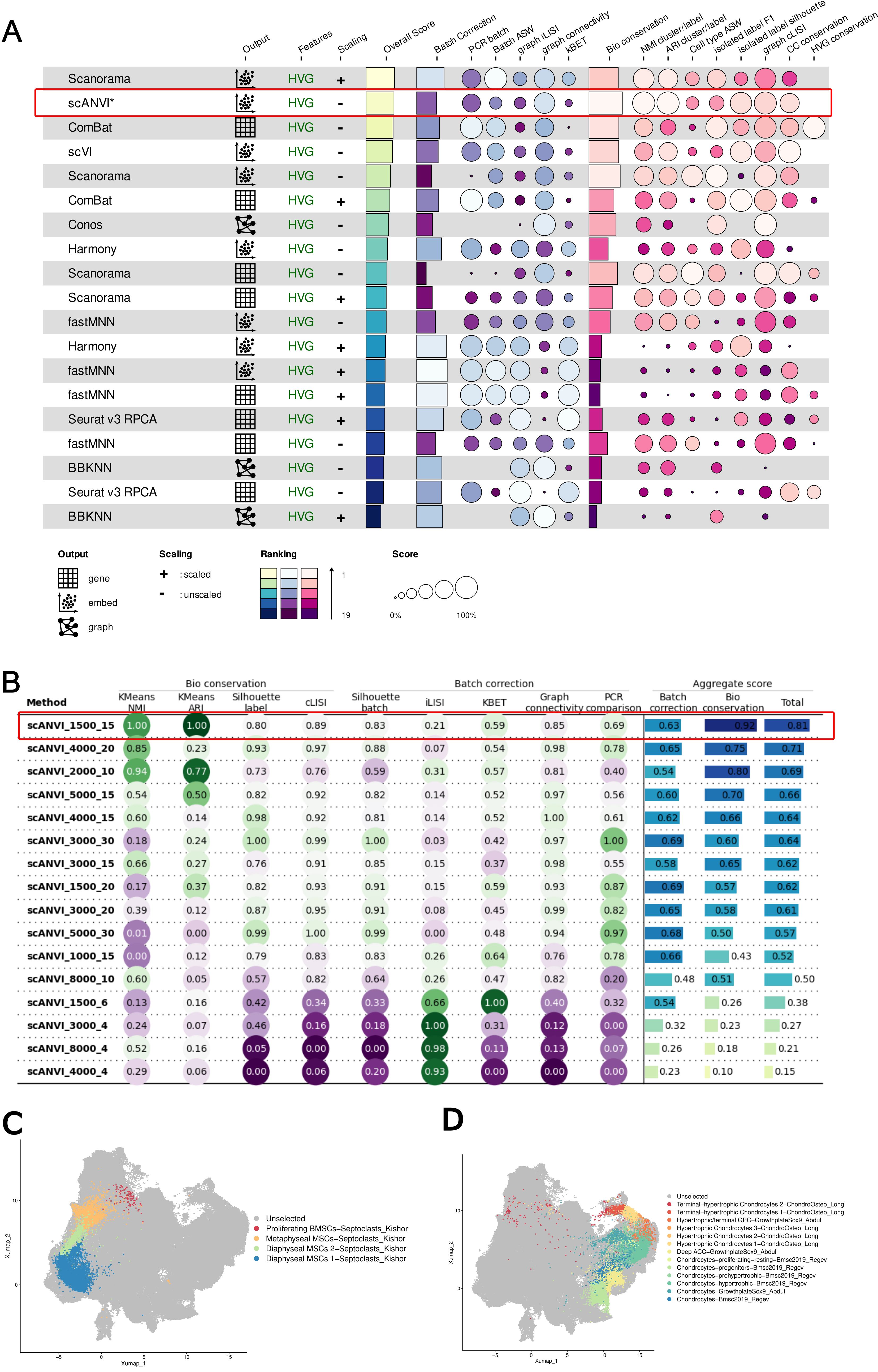
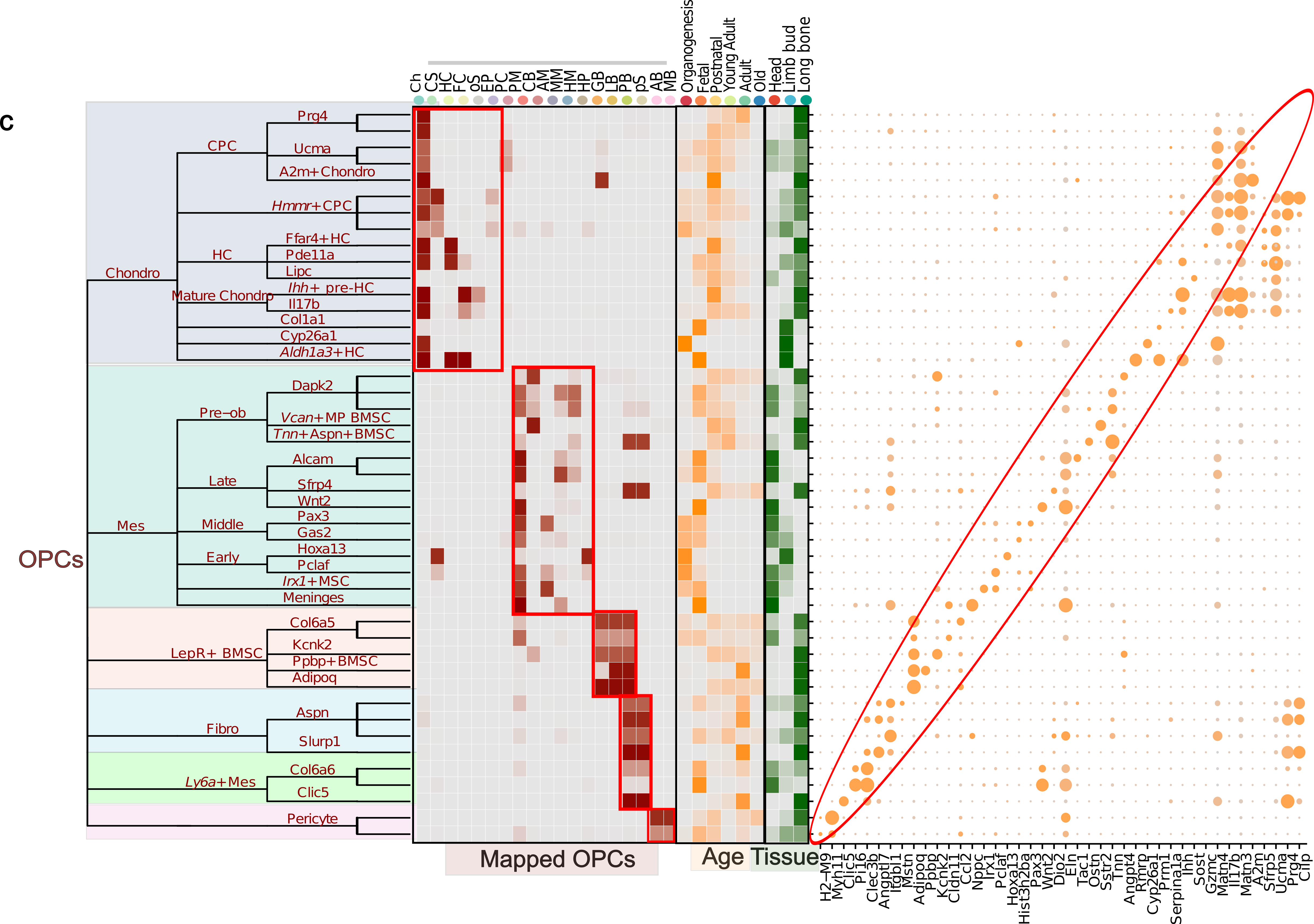
As I delved deeper into the project, I realized that I couldn’t provide arbitrary annotations. Therefore, I utilized consensus names, marker genes, tissue location, age, and experimentally-validated cell groups to define the clusters. I have a detailed discussion on this topic here.
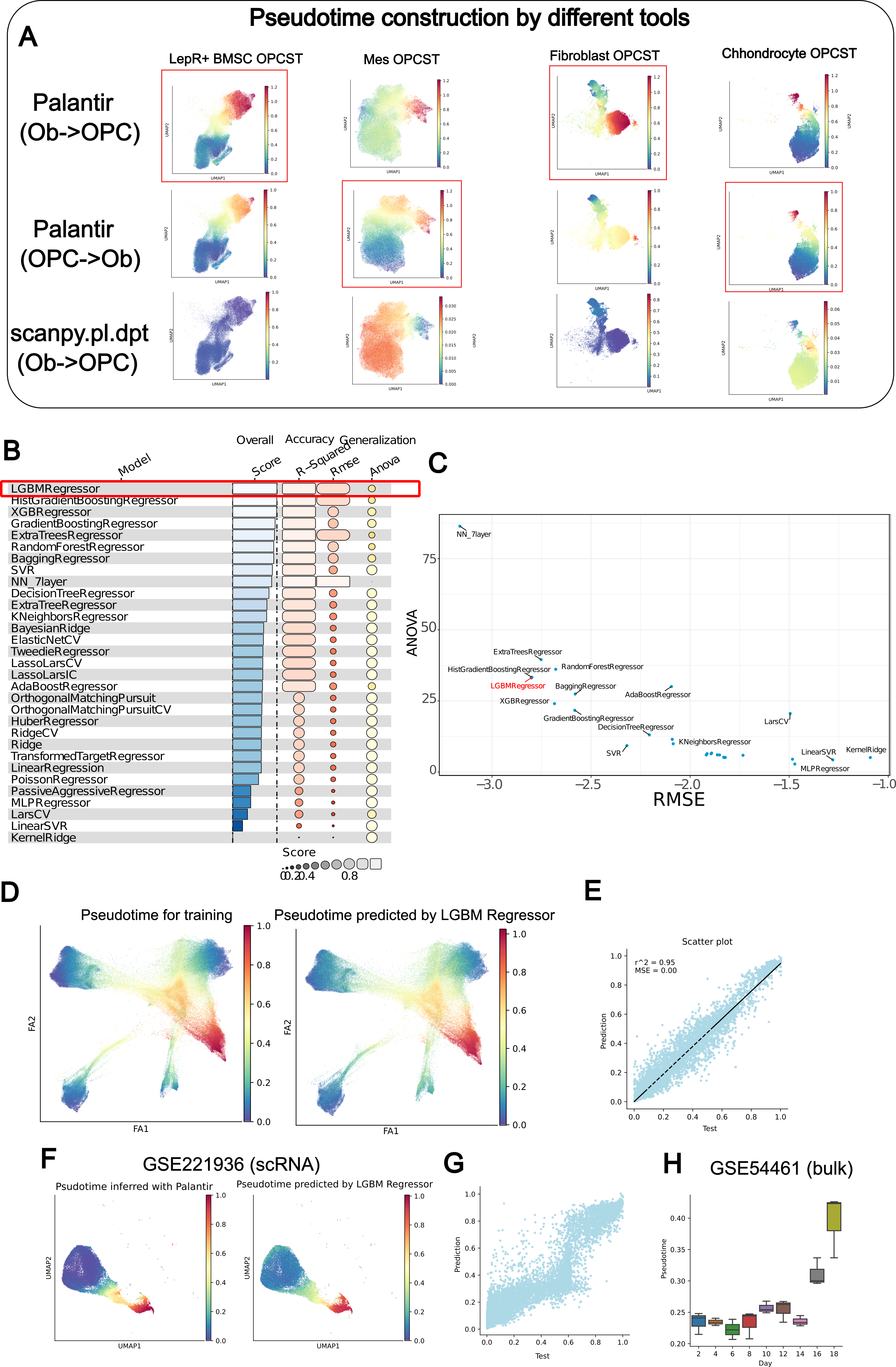
The next step is to build a model to reflect differentiation. This is complex, as there are few studies describing multiple starting points converging to a single endpoint. I discuss this problem on this page. Additionally, I spent considerable time developing a universal method to measure differentiation progression. After benchmarking various machine learning methods, I ultimately chose the LGBM Regressor for its accuracy and ability to avoid overfitting.
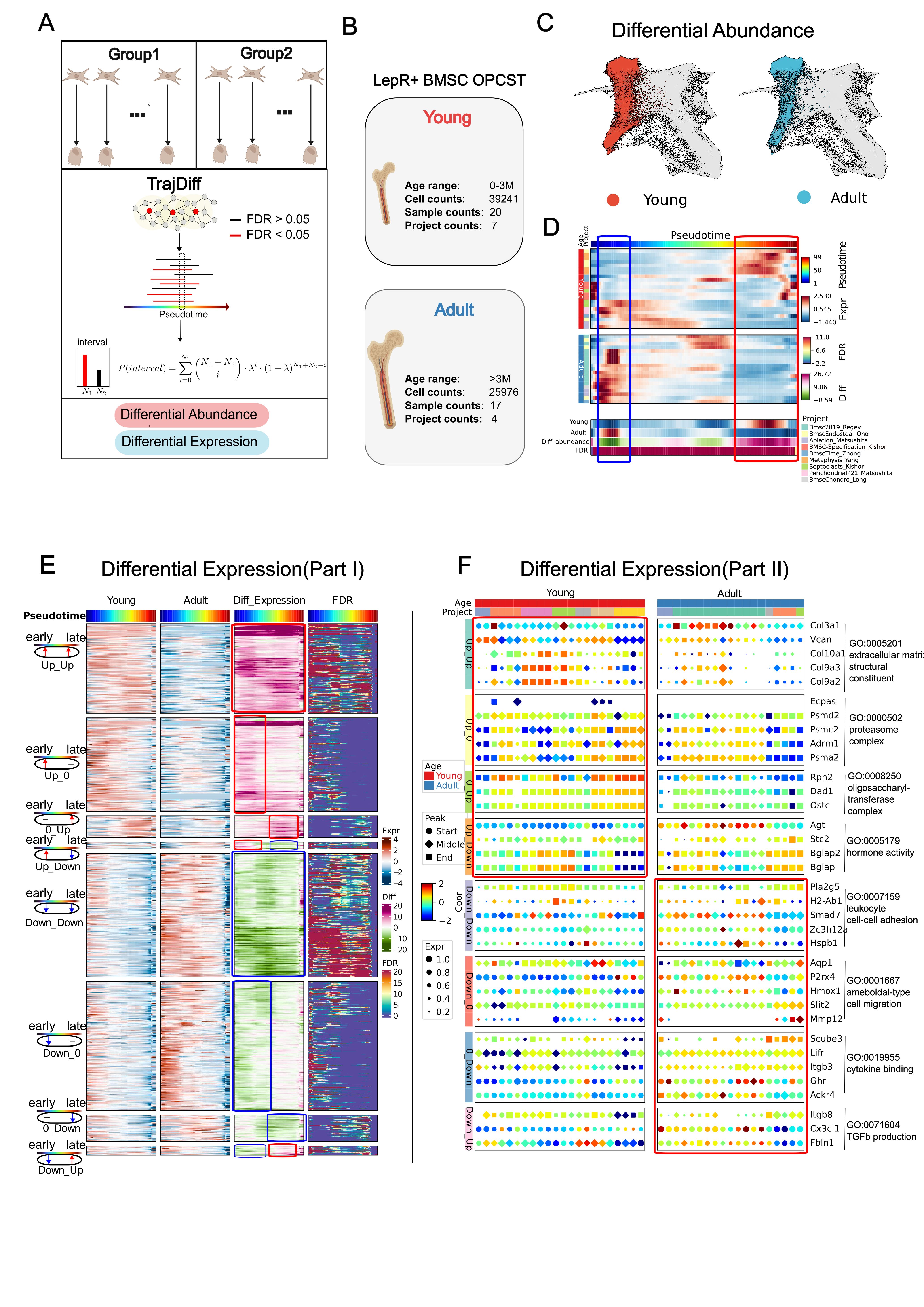
Another great idea that came to me was the ability to detect differential genes along minimal units (neighbors) of datasets. This led to the development of TrajDiff, a tool based on edgeR and binomial distribution to detect differential genes and differential abundance. During this process, Lamian and Condiments have came out. However, I believe TrajDiff is the best, as it can detect local differences and produce beautiful visualizations.
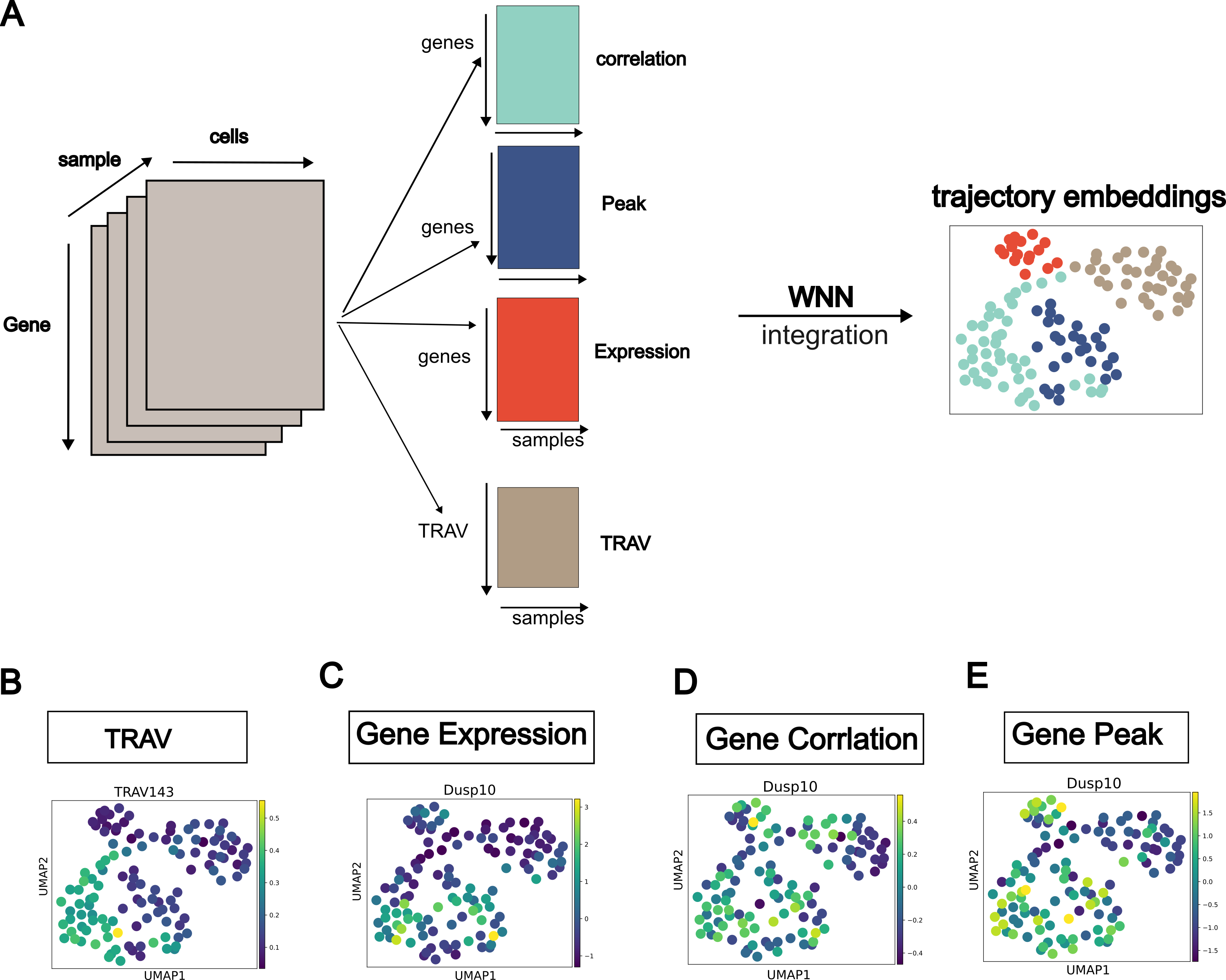
And the best part is TRAVMap. I always want to visualize the trajectory as a point, to reflect the relationship between trajectories. Ultilizing pseudotime as an axis, I found the way to connected trajectories. An additional outcome is the development of an algorithm to detect pseudotime gene modules, which was described here.
I had a very hard time combining all these elements to form a cohesive paper. I rewrote the article many times. Huan Liu provided me with a lot of help, and I found an invaluable book written by Steven Pinker, which helped me write more clearly. After submitting to PLOS Genetics, two reviewers, Noriaki Ono and Jiadong Mao, provided plenty of critical comments that greatly improved the quality of my manuscript.
Through this journey, I’ve come to realize that learning, creating, and expressing myself are some of the most beautiful experiences in my life. I’ve discovered that exploring the unknown is an essential part of my life’s purpose. This project is just a starting point, and I see it as a launchpad for what lies ahead. I am filled with anticipation for the future!