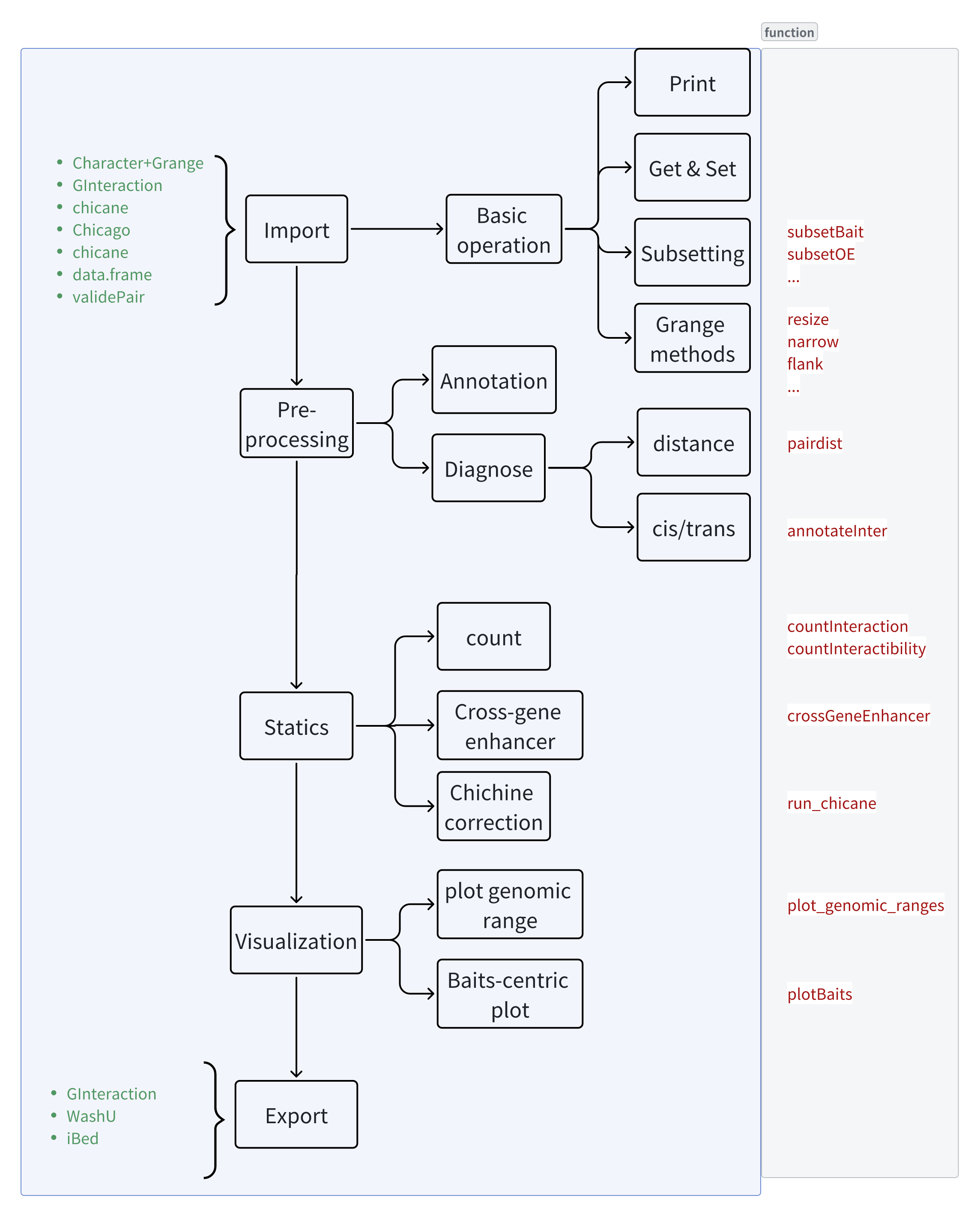
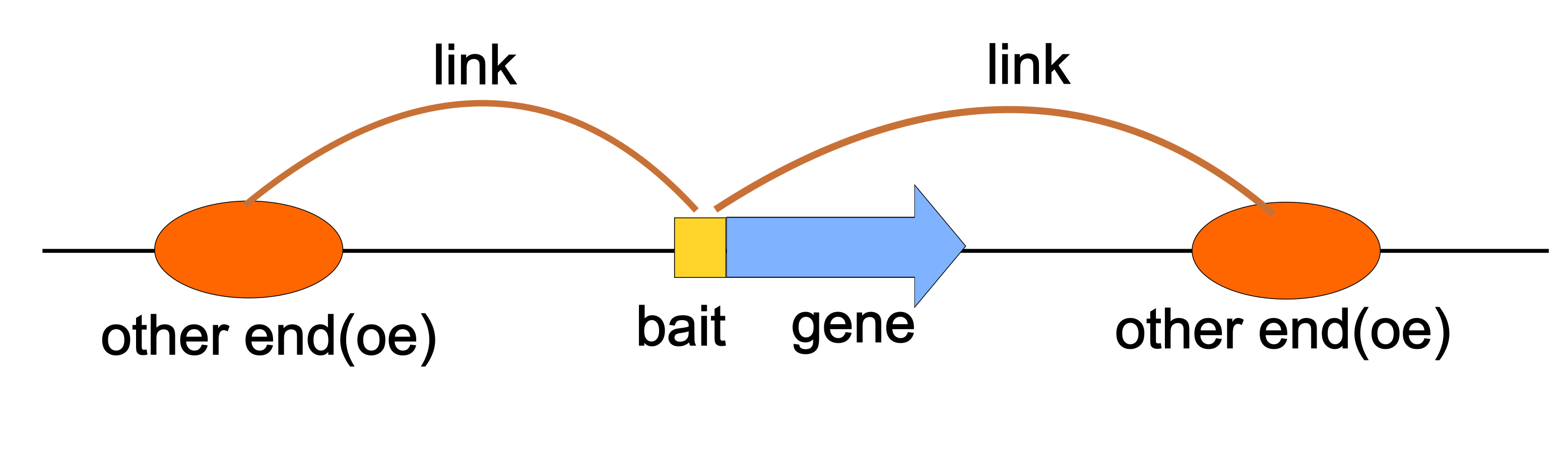
The goal of linkSet is to provide tools for working with gene-enhancer interactions, which is commonly seen in HiC, PC-HiC, and single-cell ATAC-seq data analysis.
Installation
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")linkSet is not currently available on Bioconductor. You can install it from GitHub with:
if (!requireNamespace("remotes", quietly = TRUE)) {
install.packages("remotes")
}
remotes::install_github("GilbertHan1011/linkSet")Example
LinkSet can be converted from multiple data structures, like GRanges, GInteractions, and dataframes.
suppressMessages(library(linkSet))
## basic example code
library(GenomicRanges)
gr1 <- GRanges(seqnames = c("chr1", "chr1", "chr2"),
ranges = IRanges(start = c(1, 100, 200), width = 10),
strand = "+", symbol = c("Gene1", "Gene2", "Gene3"))
gr2 <- GRanges(seqnames = c("chr1", "chr2", "chr2"),
ranges = IRanges(start = c(50, 150, 250), width = 10),
strand = "+")
## construct linkSet
ls <- linkSet(gr1, gr2, specificCol = "symbol")
ls
#> linkSet object with 3 interactions and 1 metadata column:
#> bait seqnames_oe ranges_oe | anchor1.symbol
#> <character> <Rle> <IRanges> | <character>
#> [1] Gene1 --- chr1 50-59 | Gene1
#> [2] Gene2 --- chr2 150-159 | Gene2
#> [3] Gene3 --- chr2 250-259 | Gene3
#> -------
#> regions: 6 ranges and 0 metadata columns
#> seqinfo: 2 sequences from an unspecified genome; no seqlengthsThe full manual can be reached at Documentation.